80 Best of Gatk4 Haplotypecaller Multithreading
Gatk4 haplotypecaller multithreading The haplotypecaller-gvcf-gatk4 workflow runs the HaplotypeCaller tool from GATK4 in GVCF mode on a single sample according to GATK Best Practices. Since the Spark tools are still in beta testing and.
 Pdf Recommendations For Performance Optimizations When Using Gatk3 8 And Gatk4
Pdf Recommendations For Performance Optimizations When Using Gatk3 8 And Gatk4
Overall yes these steps certainly help and theres consequences for not following them.
Gatk4 haplotypecaller multithreading. You signed in with another tab or window. The output file produced will be a. GATK4 HaplotypeCaller cant multithread like GATK3 HaplotypeCaller as GATK4 HP wont accept neither options -nct or -nt. Gatk4 haplotypecaller multithreading
BQSR will improve things and remove systematic artefacts of quality scores. In other words whenever the program encounters a region showing signs of variation it discards the existing mapping information and completely reassembles the reads in that region. This is only available on linux systems and defaults to using 4 threads. Gatk4 haplotypecaller multithreading
C Distributions of reported GQ for DeepVariant v080 in all 1248 samples computed genome-wide. D Distributions of reported GQ for GATK4 HaplotypeCaller in all 1248 samples computed on chromosome 2 only. Ill continue the investigation with people at Brown U CCV. Gatk4 haplotypecaller multithreading
You signed out in another tab or window. The HaplotypeCaller is capable of calling SNPs and indels simultaneously via local de-novo assembly of haplotypes in an active region. Its Best Practices are great guides for various analyses of sequencing data in SAMBAMCRAM and VCF formats. Gatk4 haplotypecaller multithreading
See the link above for extra detail. However the GATK was designed and primarily serves to analyze human genetic data and all its pipelines are optimized for this purpose. GATK4110---HaplotypeCaller HaplotypeCallerEngine - Disabling physical phasing which is supported only for reference-model confidence output 0 23 months ago by. Gatk4 haplotypecaller multithreading
You can read more about multithreading and parallelism in GATK here. GATK4 performs limited parallelization through multithreading that cannot be configured by the user for example by silently allocating an optimal number of threads for the variant calling step. When executed the workflow scatters the HaplotypeCaller tool over a sample using an intervals list file. Gatk4 haplotypecaller multithreading
If youre an old-timer you may be familiar with the engine arguments -nt and -nct. Note the broken y-axis and different scales. Earlier versions of the GATK used some home-grown hand-rolled code to produce multithreading. Gatk4 haplotypecaller multithreading
As you noted documentation is scattered and scarce - eg. SortSamSpark enabled 16 local cores gave rise to a speed-up of 836. The haplotypecaller-gvcf-gatk4 workflow runs the GATK4 HaplotypeCaller tool in GVCF mode on a single sample according to GATK Best Practices. Gatk4 haplotypecaller multithreading
All Spark tools can be trivially parallelized across multiple threads using the local runner and across a cluster using spark-submit or gcloud. The optimal PGC threads number is 2 for GATK4 MarkDuplicates. When executed the workflow scatters the HaplotypeCaller tool over the input bam sample using an interval list file. Gatk4 haplotypecaller multithreading
To specify the number of threads you wish to use with HaplotypeCaller include --native-pair-hmm-threads documentation. So this step is optional. In other words whenever the program encounters a region showing signs of variation it discards the existing mapping information and completely reassembles the reads in that region. Gatk4 haplotypecaller multithreading
B Genotype quality calibration for GATK4 HaplotypeCaller analogous to A. Thus we recommend GATK4 running multiple samples on one node. To refresh your session. Gatk4 haplotypecaller multithreading
The multithreading version of GATK4 HaplotypeCaller is actually HaplotypeCallerSpark as spark args arent taken by GATK4 HP. One specific one is the option to use multiple threads with HaplotypeCallers pairHmm. Optimal threads number is 12 for GATK4 HaplotypeCaller in ERC mode giving rise to 124 speed-up. Gatk4 haplotypecaller multithreading
Reload to refresh your session. How to Run Spark-enabled GATK tools on a local multi-core machine. The HaplotypeCaller is capable of calling SNPs and indels simultaneously via local de-novo assembly of haplotypes in an active region. Gatk4 haplotypecaller multithreading
Reload to refresh your session. This will only parallelize the pair hidden Markov models pair HMM process. The latest versions of GATK GATK4 contains Spark and traditional implementations that is the Walker mode which improve runtime performance dramatically from previous versions. Gatk4 haplotypecaller multithreading
There are few limited multithreaded options remaining in GATK4 outside of spark. Based on this Spark GATK4 page you can try. The total walltime will be 341 hours on 40 samples with 118 samples processed per hour at the cost of 260 per sample on. Gatk4 haplotypecaller multithreading
In GATK4 we take advantage instead of an open-source industry-standard software library called Spark produced by the Apache Software Foundation. In GATK4 the way to make a tool multithreaded is to implement it as a Spark tool. And multi-threading with Spark local runner highly speeded up GATK4 tool execution. Gatk4 haplotypecaller multithreading
The GATK Genome Analysis Toolkit is the most used software for genotype calling in high-throughput sequencing data in various organisms. See if I understood. Updated In a nutshell Spark is a piece of software that GATK4 uses to do multithreading which is a form of parallelization that allows a computer or cluster of computers to finish executing a task sooner. Gatk4 haplotypecaller multithreading
The HaplotypeCaller will perform graph based assembly which should fix issues that the UnifiedGenotyper would miss. Some users parallelize the variant calling step by splitting the input BAM file by chromosome to call the variants per chromosome and later merge the. GATK4 best practice pipelines published by Broad Institute2 are widely adopted by the genomics community. Gatk4 haplotypecaller multithreading
This is an argument for the method HaplotypeCaller itself so it should be located after the method is called as opposed to one of the --java-options. In GATK4 multithreading is implemented using Spark see Document how multi-threading support works in GATK4. Gatk4 haplotypecaller multithreading
Https Www Biorxiv Org Content 10 1101 2020 02 10 942086v2 Full Pdf
Plos One Elprep 4 A Multithreaded Framework For Sequence Analysis
Https Www Biorxiv Org Content 10 1101 2020 05 17 101105v1 Full Pdf
Https Www Biorxiv Org Content 10 1101 348565v1 Full Pdf
Https Biohpc Cornell Edu Lab Doc Variant Workshop Pdf
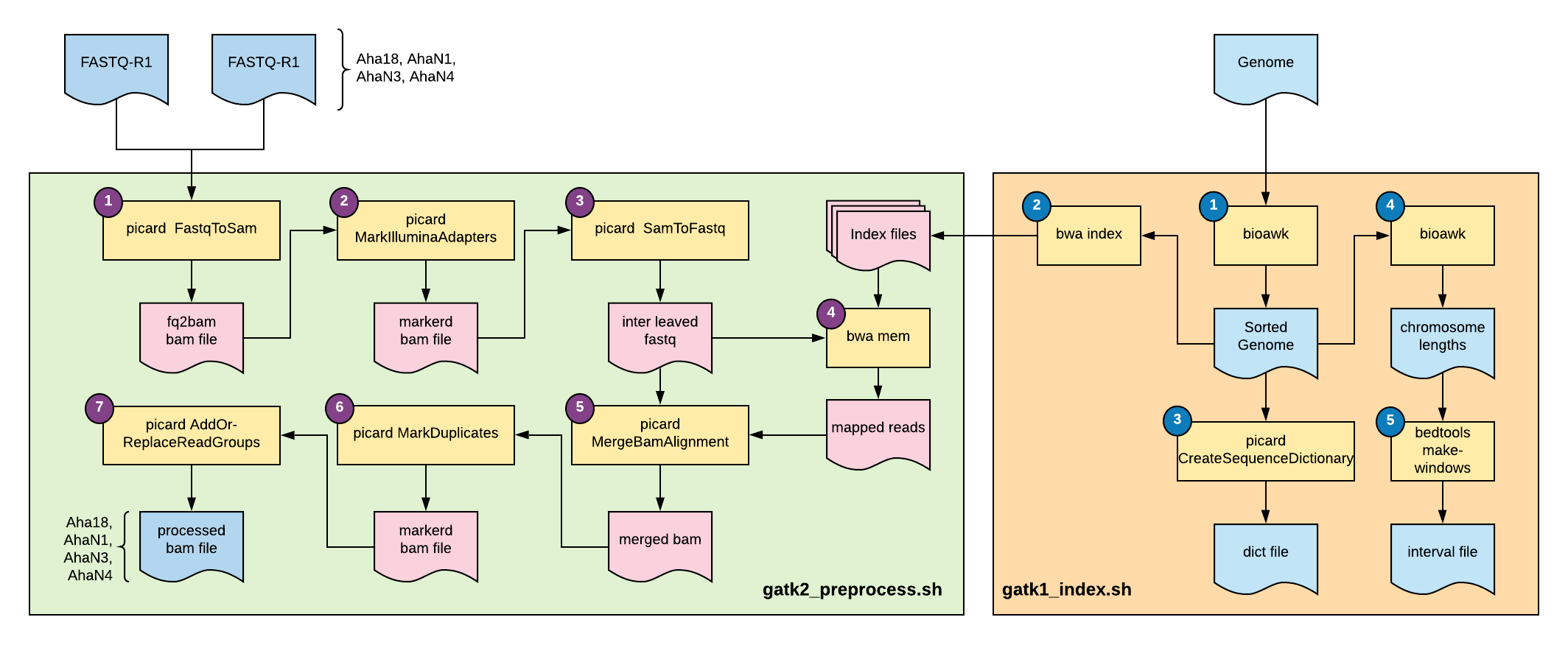 Gatk Best Practices Workflow For Dna Seq Bioinformatics Workbook
Gatk Best Practices Workflow For Dna Seq Bioinformatics Workbook
Https Bmcbioinformatics Biomedcentral Com Track Pdf 10 1186 S12859 019 3169 7
 Multithreaded Variant Calling In Elprep 5 Biorxiv
Multithreaded Variant Calling In Elprep 5 Biorxiv
Https Repository Tudelft Nl Islandora Object Uuid 7d02ec4a 0d99 453a 8950 F54287d91e2a Datastream Obj Download
Multithreaded Variant Calling In Elprep 5
Plos One Multithreaded Variant Calling In Elprep 5
 Wgs Benchmarks Runtime Ram Use And Disk Use In Gatk 4 Vs Elprep 4 Download Scientific Diagram
Wgs Benchmarks Runtime Ram Use And Disk Use In Gatk 4 Vs Elprep 4 Download Scientific Diagram
Https Trepo Tuni Fi Bitstream Handle 123456789 27612 Luo Pdf Sequence 4 Isallowed Y
Https Academic Oup Com Bioinformatics Advance Article Pdf Doi 10 1093 Bioinformatics Btaa1081 35888806 Btaa1081 Pdf
 Elprep 4 A Multithreaded Framework For Sequence Analysis Biorxiv
Elprep 4 A Multithreaded Framework For Sequence Analysis Biorxiv
 Accurate Scalable Cohort Variant Calls Using Deepvariant And Glnexus Biorxiv
Accurate Scalable Cohort Variant Calls Using Deepvariant And Glnexus Biorxiv
 Wes Benchmarks Runtime Ram Use And Disk Use In Gatk 4 Vs Elprep 4 Download Scientific Diagram
Wes Benchmarks Runtime Ram Use And Disk Use In Gatk 4 Vs Elprep 4 Download Scientific Diagram
Https Www Ibm Com Downloads Cas Zjqd0qal

